Action 5 – Assessment of inbreeding and genetic diversity in populations
Action 5 aims to calculate inbreeding and genetic diversity in 26 breeds of 4 poultry species. Genotypes obtained by both TuBAvI and TuBAvI-2 will be merged in a dataset with a total of more than 1000 birds and results will be analysed. developed to reduce inbreeding in populations with excess of homozygosity.
Inbreeding, genetic diversity and the risk of extinction will be evaluated in populations characterised with microsatellite markers. TuBAvI and TuBAvI-2 datasets will be integrated to calculate the contribution to biodiversity and estimate the genetic distance among breeds.
Finally, genetic diversity among breeds will be studied with respect to gene PAX7 polymorphisms.
To see the results from the first TuBAvI project, click here.
Action 5 – TuBAvI (2017-2020): results
Evaluation of inbreeding, genetic diversity and contribution of each breed to biodiversity (UniTO)
Genotypes obtained in Action 2 were analysed for the evaluation of inbreeding (mean H0 per breed), genetic diversity, and contribution of each breed to biodiversity. Extinction risk index and population structure were studied in each breed.
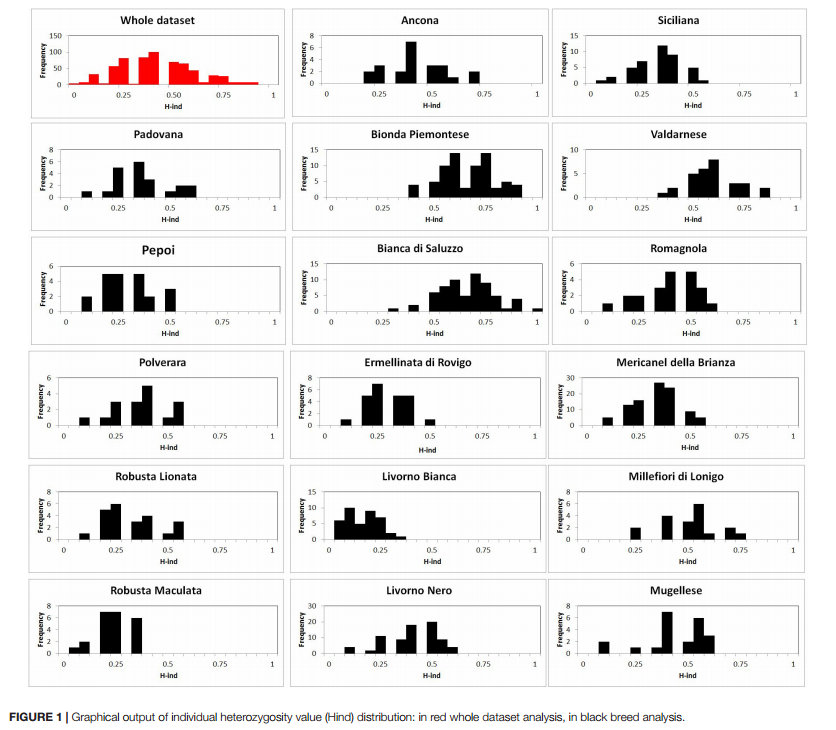
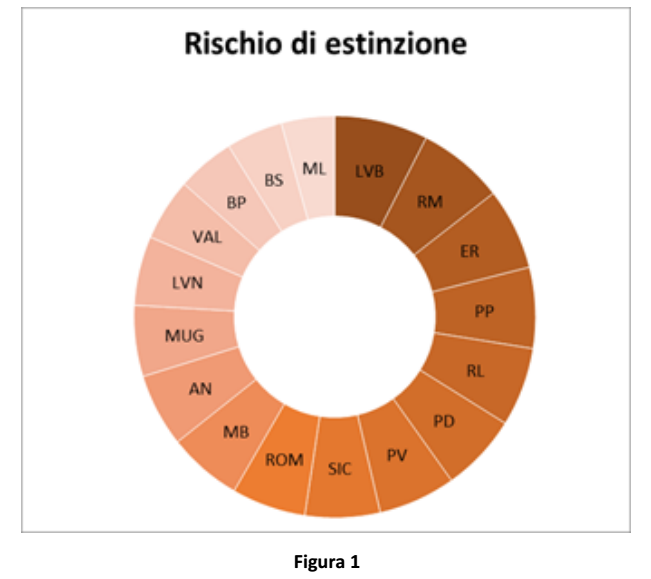
Distribution of individual molecular inbreeding (Average individual heterozygosity, H-ind) in the whole population (red bars) and in single populations (black bars) is shown in Figure 1. Overall distribution shows a bell-like shape, with 60% inbreeding in most birds.
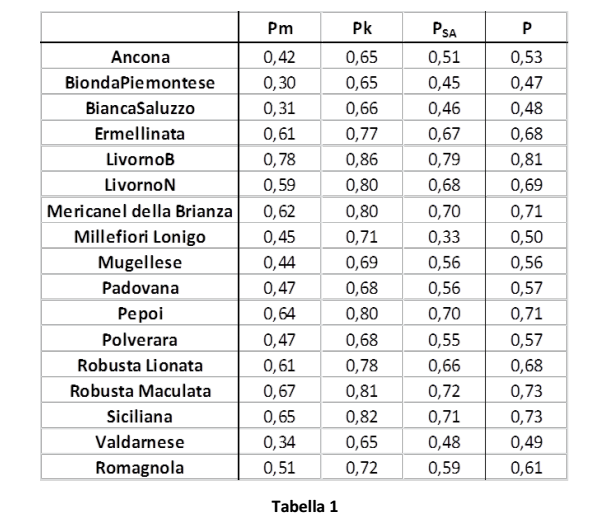
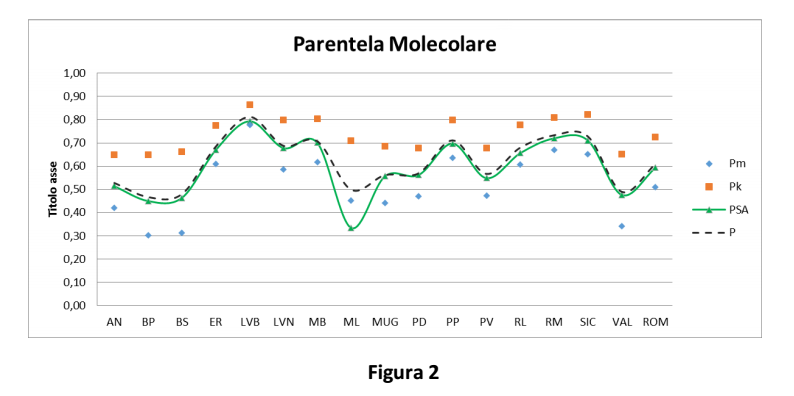
Molecular kinship in the populations (Table 1 and Figure 2) was calculated as the average of each individual’s kinship and compared to all the other individuals from the same population:
Pm: molecular kinship calculated according to Molkin (Caballero and Toro, 2002; Eding and Meuwissen, 2001)
Pk: kinship calculated on the basis of Kinship Distances (Caballero and Toro, 2002)
PSA: kinship calculated on the basis of the number of shared alleles
P: average of kinship values (Pm, Pk, and PSA)
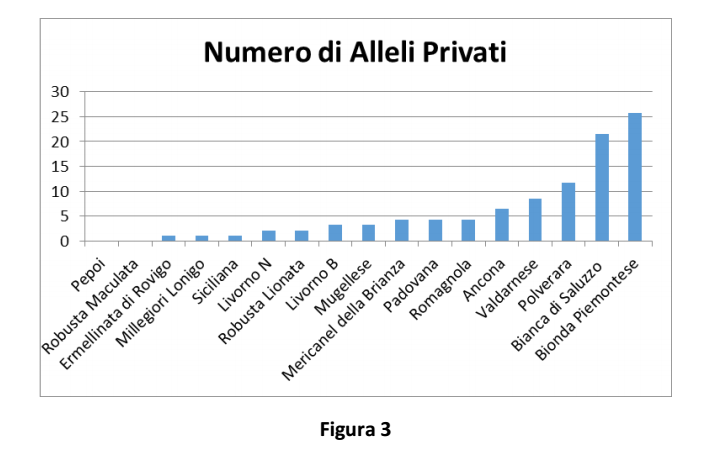
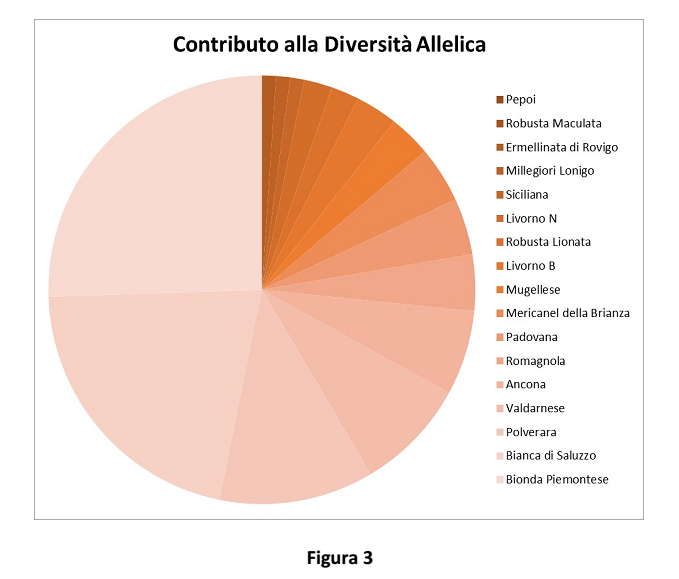
The number of private alleles shown in Figure 3 represents a peculiar breed index.
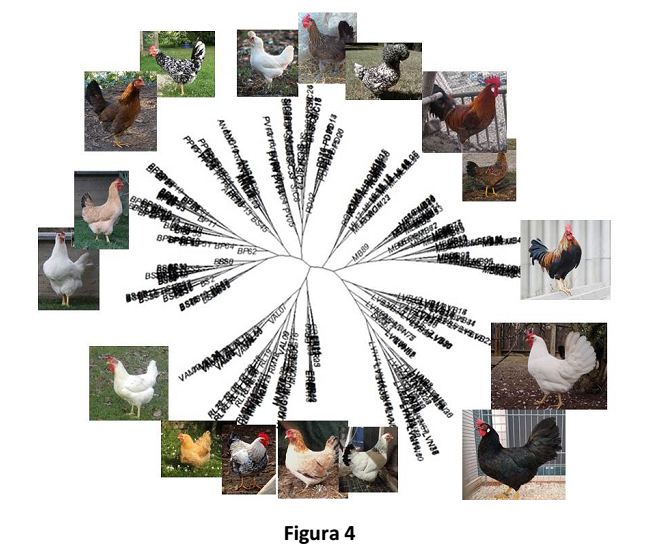
The graphic representation of genetic distances (DSH) of individuals belonging to different breeds is shown in Figure 4. Each breed’s cluster can be clearly distinguished.
Extinction Risk Index (IRC) integrates the other calculated indexes. In Figure 1, darker colours mean higher extinction risk.
The contribution of each breed to genetic variability of the species, as a contribution to biodiversity, was evaluated both as a contribution to overall genetic diversity (Figure 2), according to the method suggested by Caballero and Toro in 2002, and as a contribution to allelic richness (Figure 3).